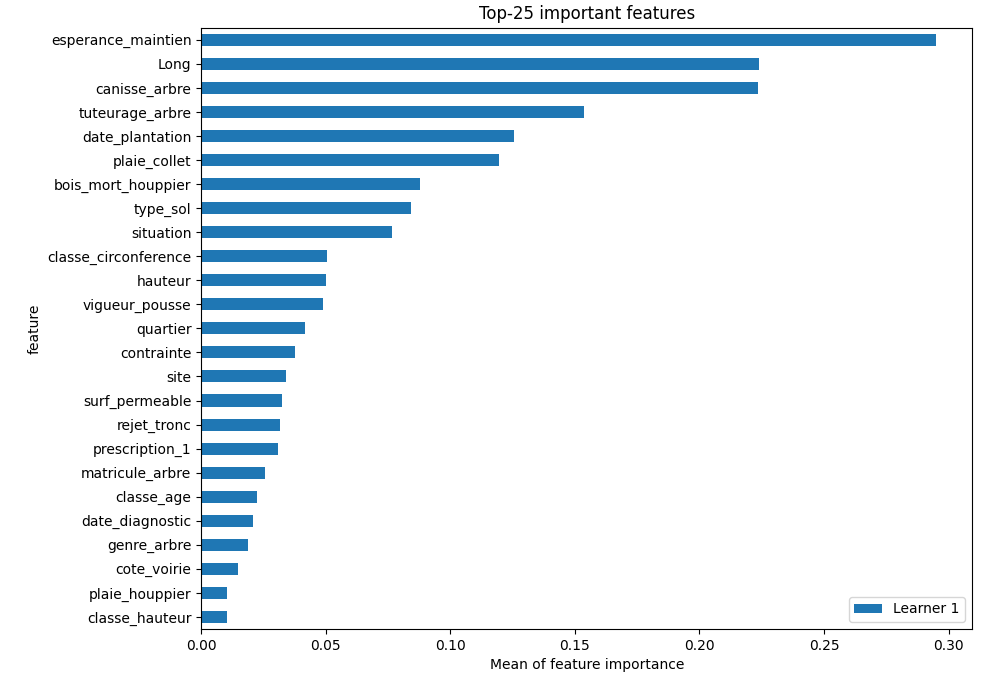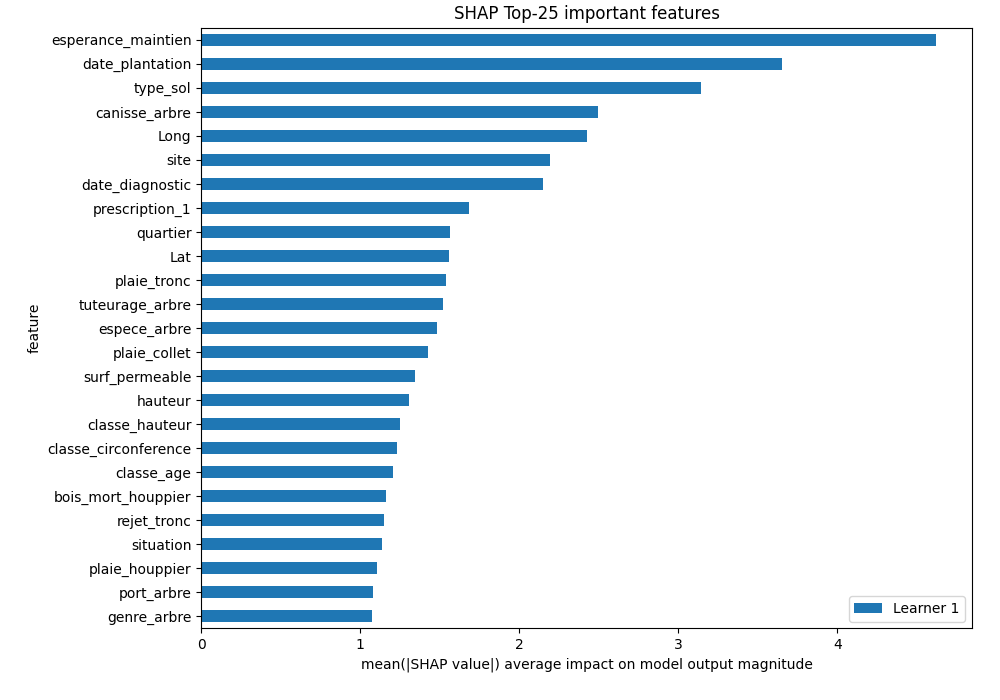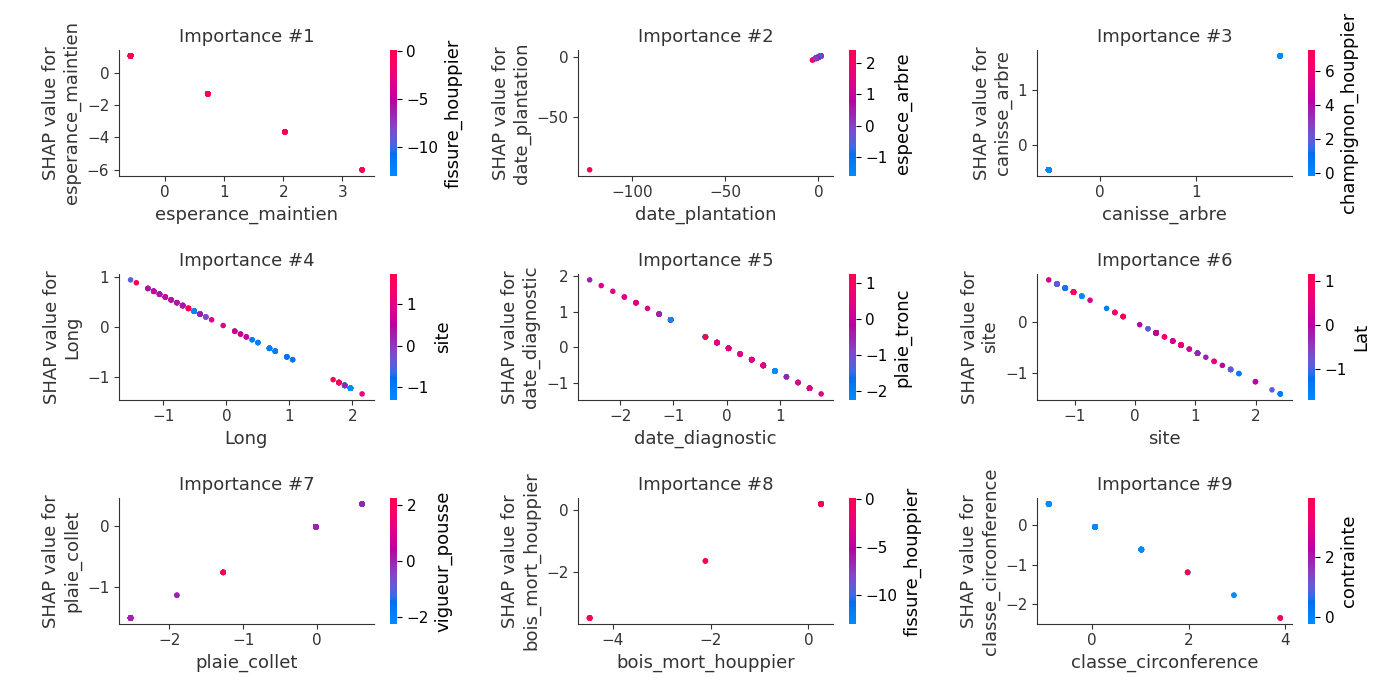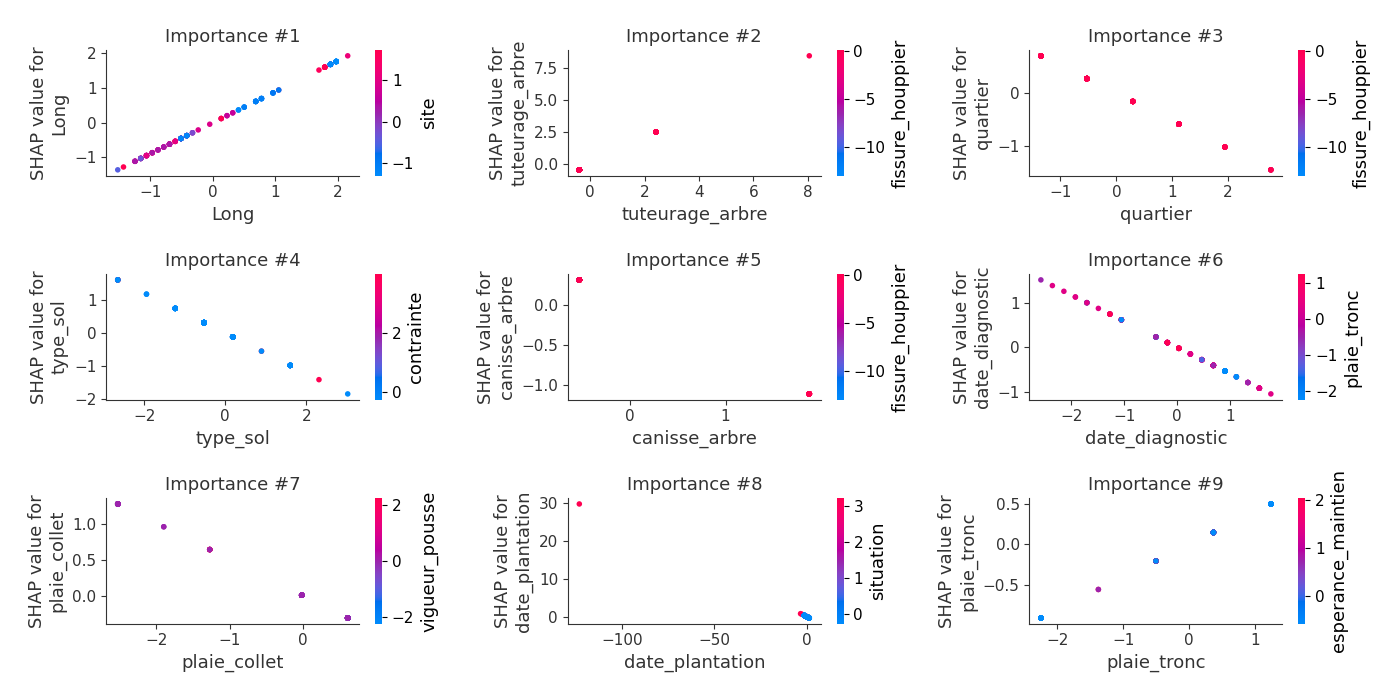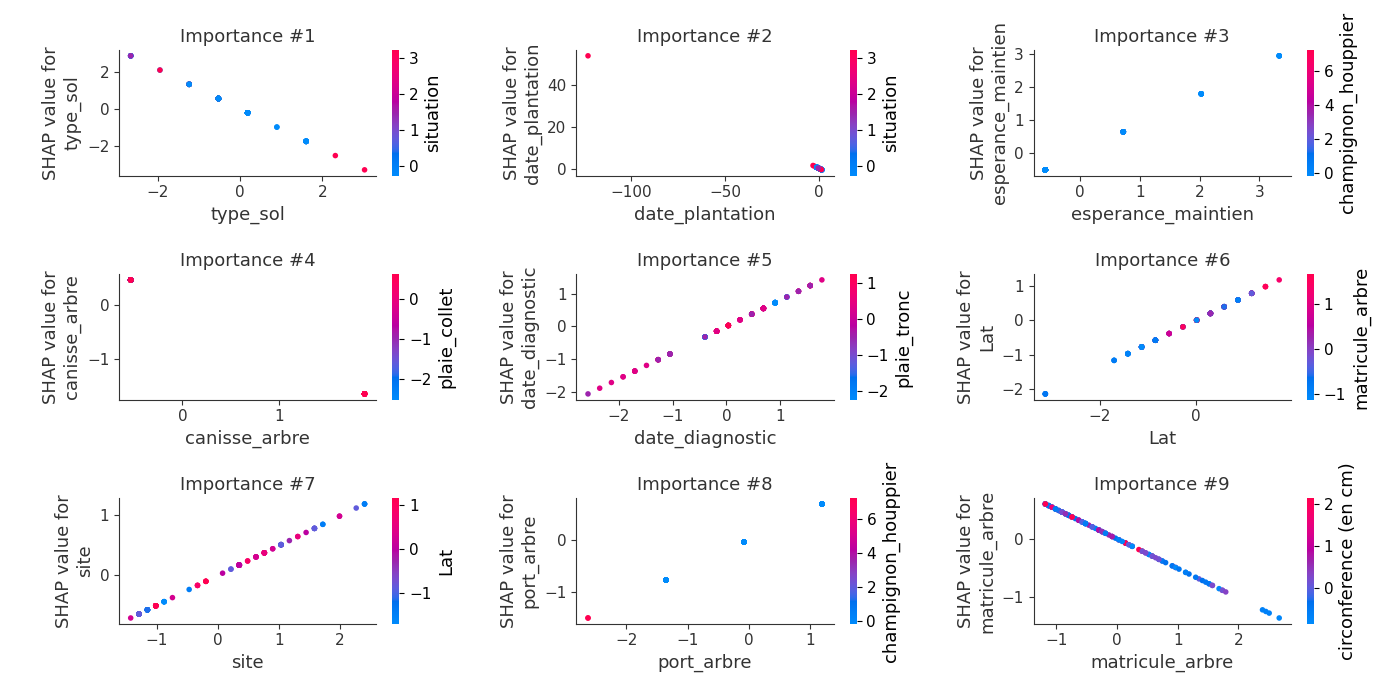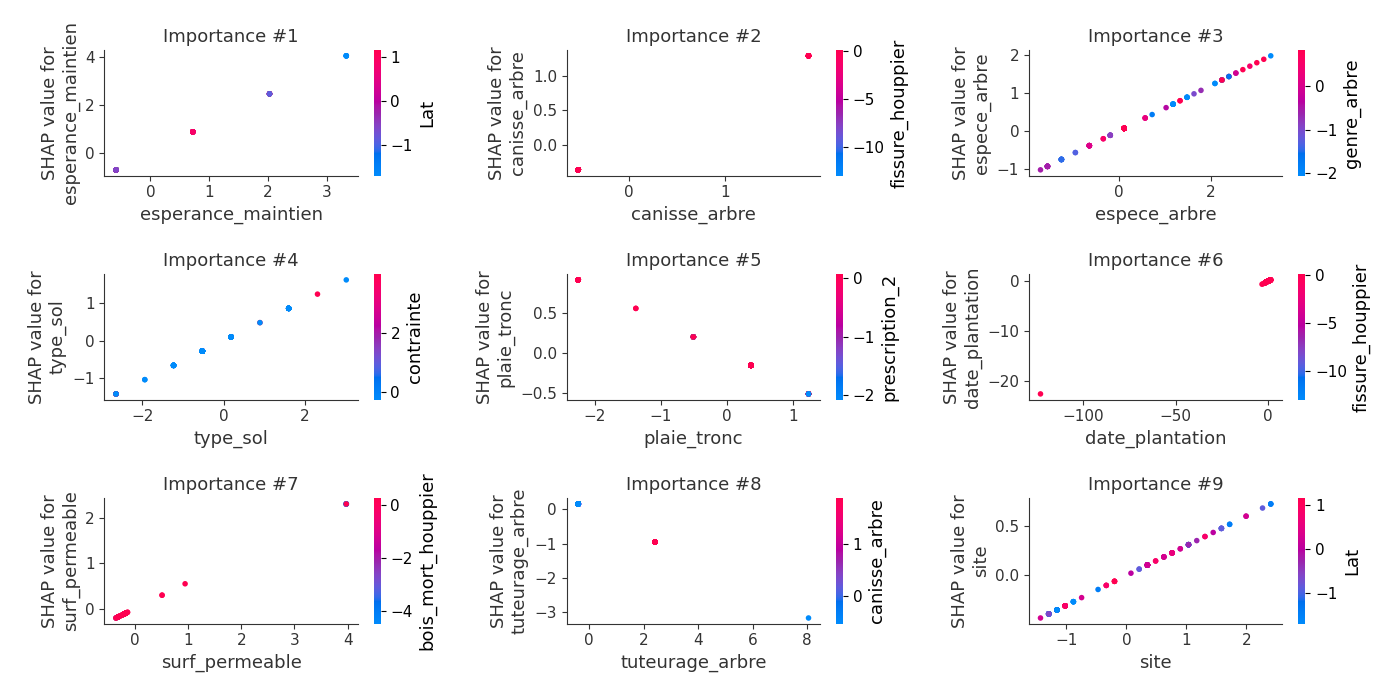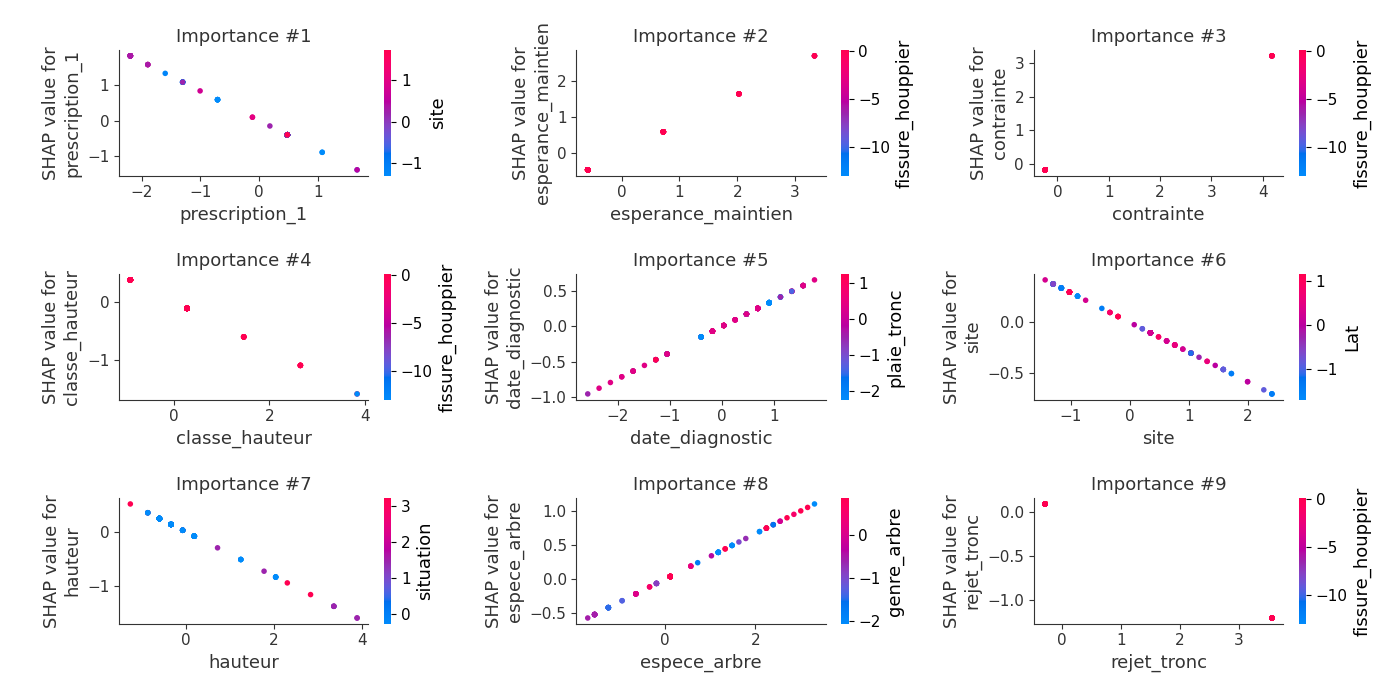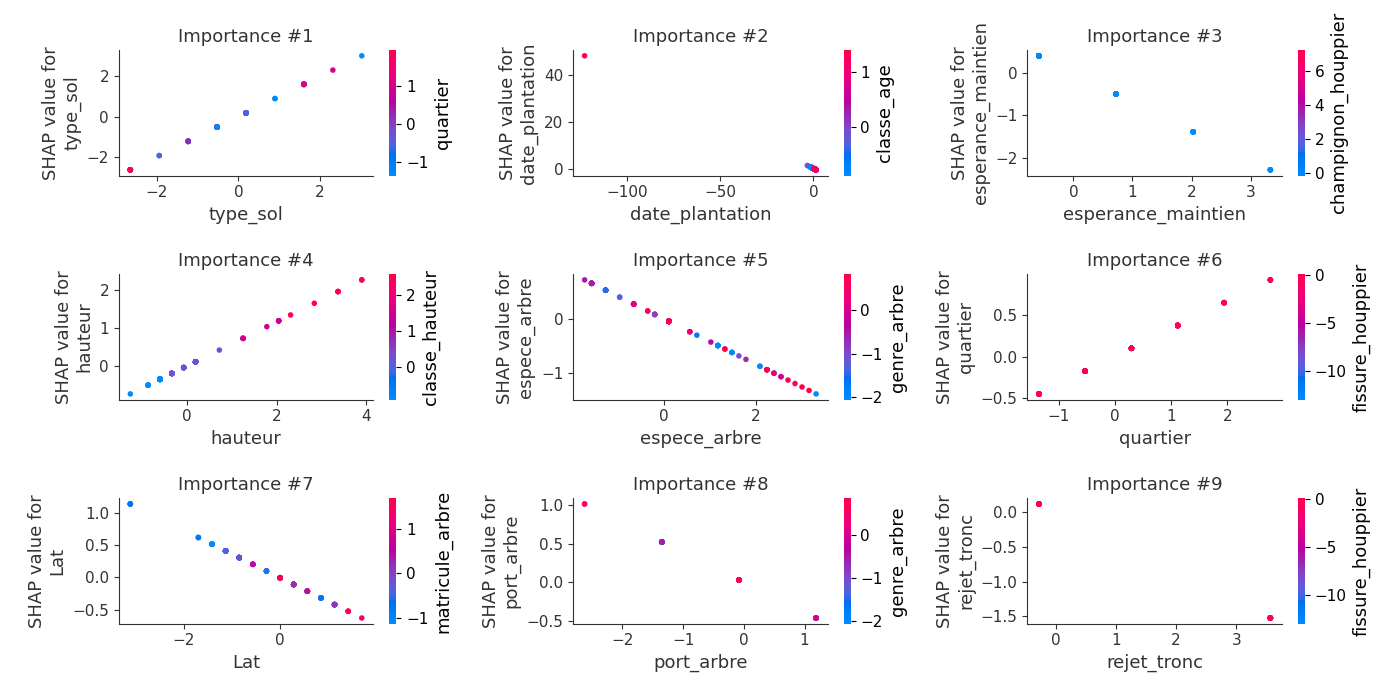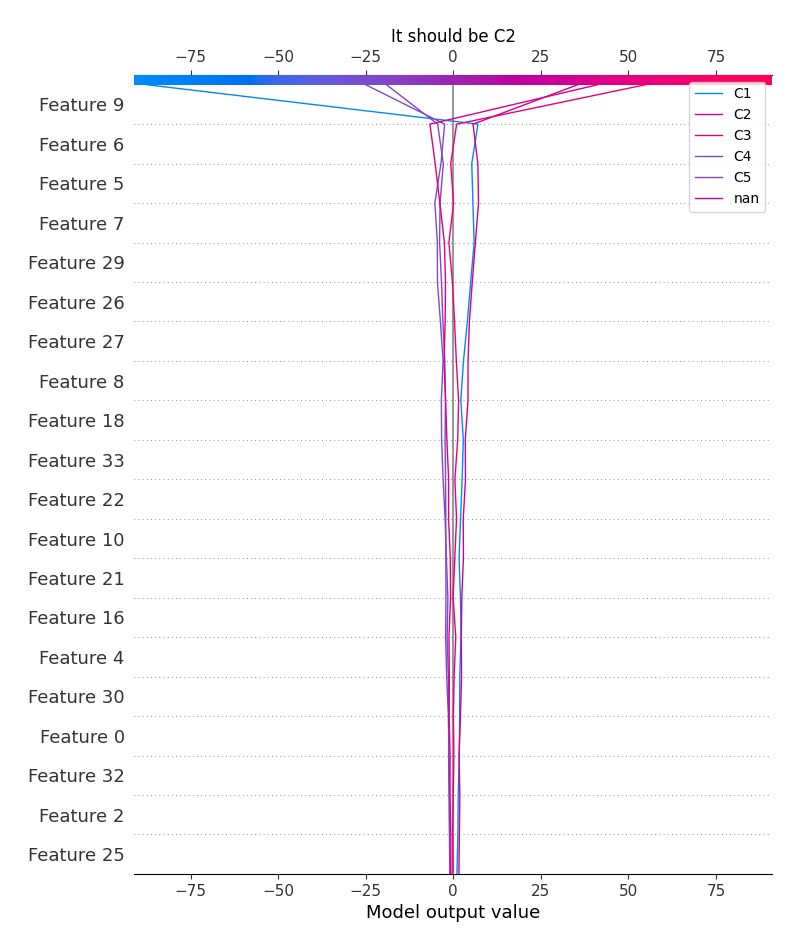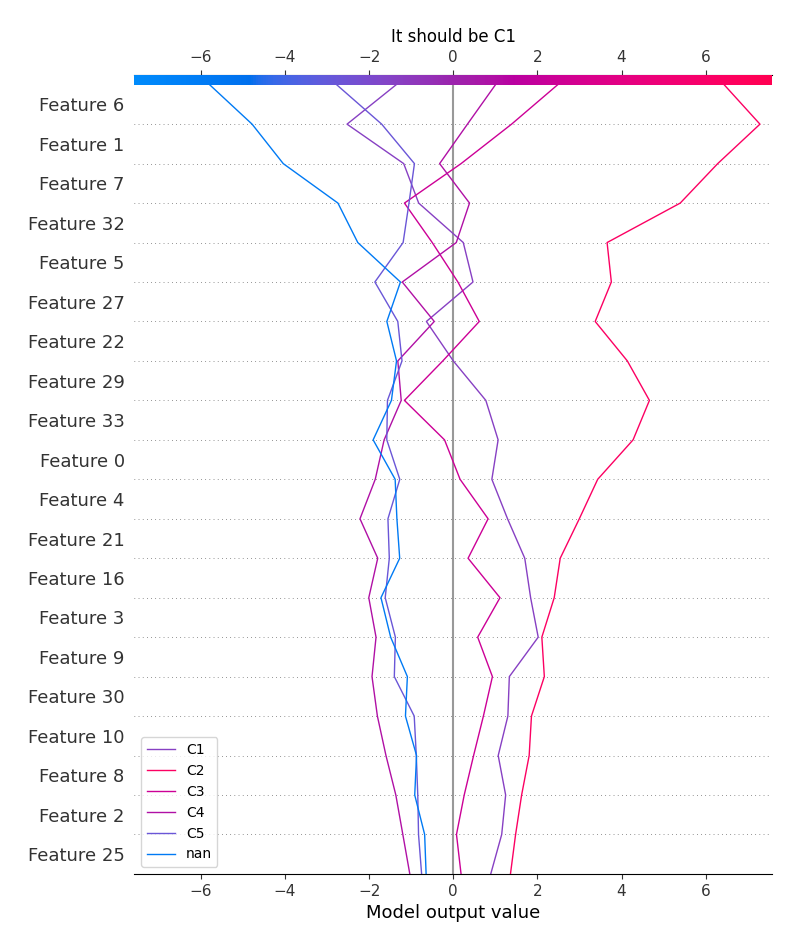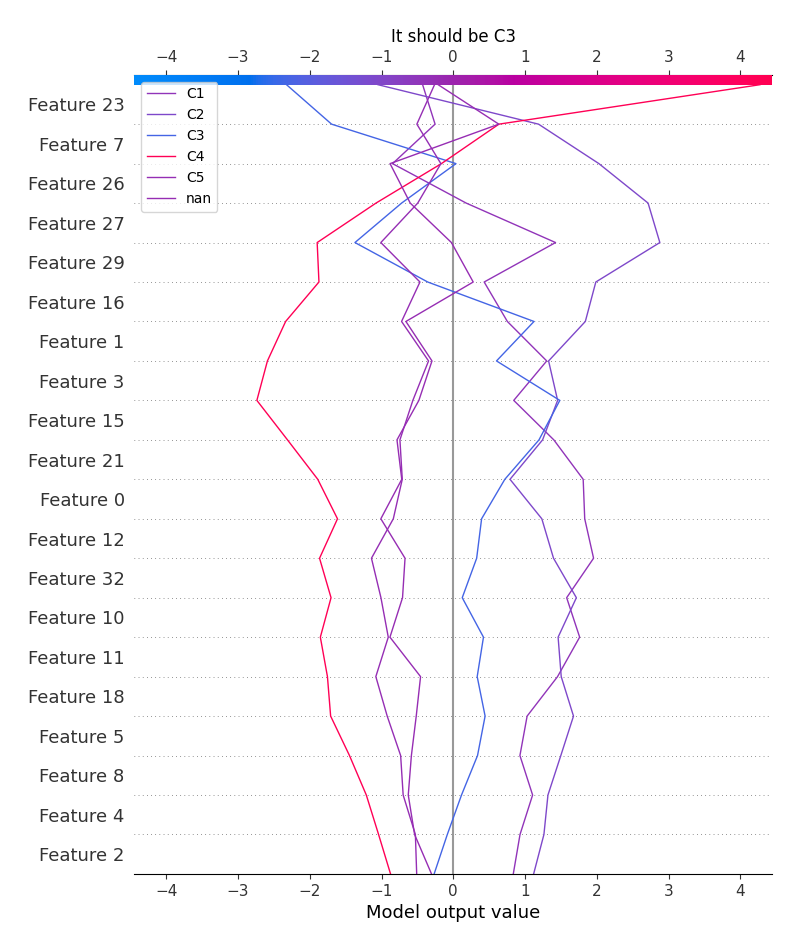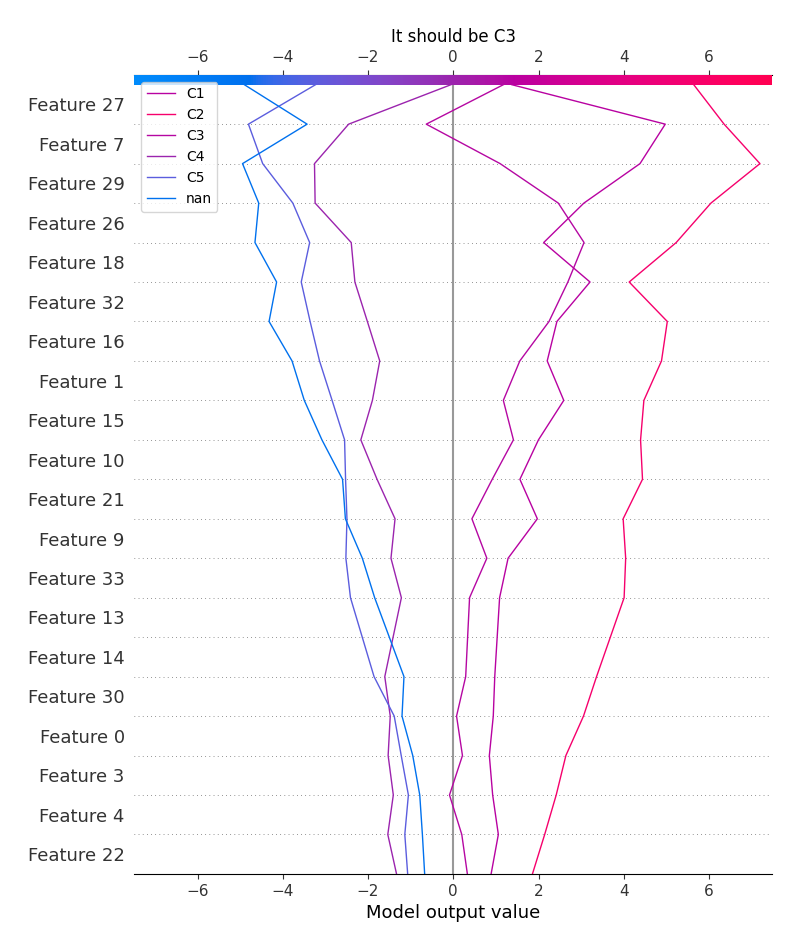
logloss
14.0 seconds
| C1 | C2 | C3 | C4 | C5 | nan | accuracy | macro avg | weighted avg | logloss | |
|---|---|---|---|---|---|---|---|---|---|---|
| precision | 0.825 | 0.848101 | 0.666667 | 0.833333 | 1 | 0.833333 | 0.838028 | 0.834406 | 0.83647 | 0.515071 |
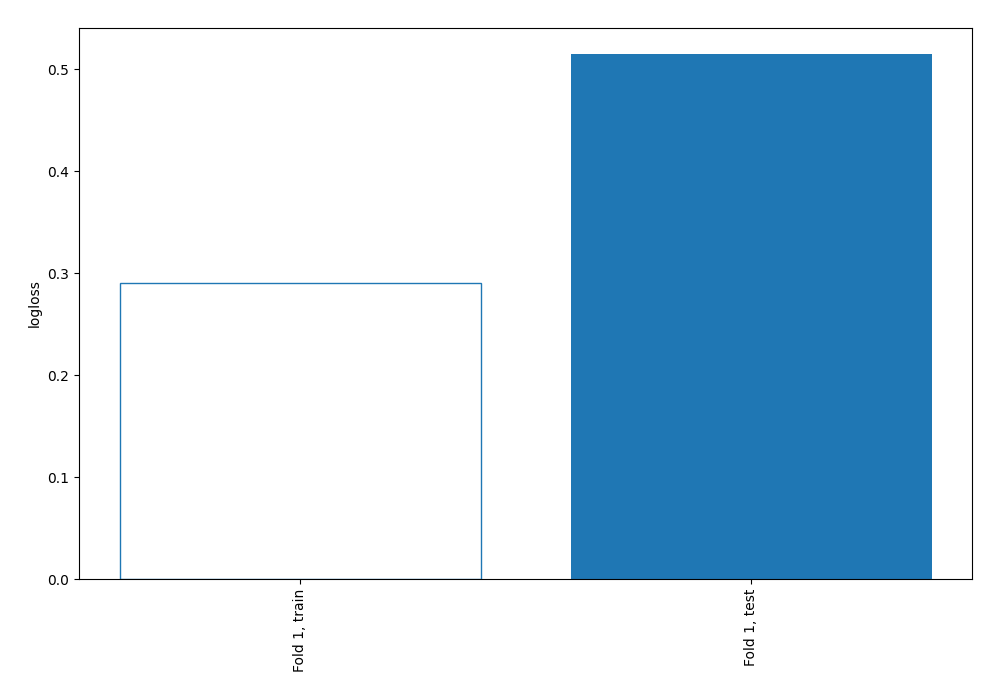
| recall | 0.767442 | 0.87013 | 0.571429 | 1 | 1 | 1 | 0.838028 | 0.868167 | 0.838028 | 0.515071 |
| f1-score | 0.795181 | 0.858974 | 0.615385 | 0.909091 | 1 | 0.909091 | 0.838028 | 0.847954 | 0.836144 | 0.515071 |
| support | 43 | 77 | 7 | 5 | 5 | 5 | 0.838028 | 142 | 142 | 0.515071 |
| Predicted as C1 | Predicted as C2 | Predicted as C3 | Predicted as C4 | Predicted as C5 | Predicted as nan | |
|---|---|---|---|---|---|---|
| Labeled as C1 | 33 | 10 | 0 | 0 | 0 | 0 |
| Labeled as C2 | 7 | 67 | 2 | 0 | 0 | 1 |
| Labeled as C3 | 0 | 2 | 4 | 1 | 0 | 0 |
| Labeled as C4 | 0 | 0 | 0 | 5 | 0 | 0 |
| Labeled as C5 | 0 | 0 | 0 | 0 | 5 | 0 |
| Labeled as nan | 0 | 0 | 0 | 0 | 0 | 5 |

| C1 | C2 | C3 | C4 | C5 | nan | |
|---|---|---|---|---|---|---|
| intercept | 1.89661 | 4.73311 | 0.313168 | -1.85347 | -2.43957 | -2.64985 |
| quartier | -0.0690232 | -0.520492 | 0.280769 | -0.197011 | 0.175582 | 0.330176 |
| site | -0.582602 | 0.359342 | 0.494725 | 0.297185 | -0.293456 | -0.275193 |
| cote_voirie | -0.057919 | -0.00634577 | -0.265585 | 0.170897 | -0.0815646 | 0.240517 |
| matricule_arbre | 0.232195 | -0.152501 | -0.5127 | 0.115912 | 0.167134 | 0.14996 |
| genre_arbre | 0.224655 | -0.155951 | 0.354036 | -0.218611 | -0.185036 | -0.0190935 |
| espece_arbre | -0.127532 | -0.106983 | -0.275406 | 0.595506 | 0.330809 | -0.416395 |
| situation | 0.347032 | -0.308958 | 0.339237 | 0.22492 | -0.31169 | -0.290541 |
| type_sol | 0.334279 | -0.610421 | -1.08511 | 0.532669 | -0.163595 | 0.992174 |
| surf_permeable | 0.749115 | -0.129183 | -0.689116 | 0.578937 | -0.118665 | -0.391089 |
| date_plantation | 0.770019 | -0.242307 | -0.438837 | 0.182992 | 0.118448 | -0.390315 |
| classe_age | 0.269413 | -0.131004 | 0.346802 | -0.231565 | 0.0344866 | -0.288132 |
| hauteur | -0.422751 | 0.30564 | -0.130745 | 0.0744087 | -0.41052 | 0.583967 |
| classe_hauteur | 0.197638 | 0.333466 | -0.0657908 | -0.335988 | -0.415089 | 0.285764 |
| diametre | -0.00137278 | -0.249939 | -0.0491795 | -0.338085 | 0.277887 | 0.360689 |
| circonference (en cm) | -0.00137278 | -0.249939 | -0.0491795 | -0.338085 | 0.277887 | 0.360689 |
| classe_circonference | -0.600672 | 0.0687286 | 0.274753 | -0.363808 | 0.246348 | 0.37465 |
| port_arbre | 0.14454 | -0.00263849 | 0.568355 | -0.197853 | -0.125059 | -0.387345 |
| vigueur_pousse | 0.778623 | -0.25484 | 0.201818 | 0.101664 | -0.493926 | -0.333338 |
| plaie_collet | 0.602204 | -0.50791 | -0.195289 | 0.0165047 | -0.141935 | 0.226425 |
| rejet_tronc | 0.305942 | -0.0402351 | 0.575186 | -0.0749786 | -0.33891 | -0.427004 |
| tuteurage_arbre | -0.211183 | 1.05375 | -0.328805 | -0.393688 | -0.109677 | -0.0103937 |
| canisse_arbre | 0.872519 | -0.590594 | -0.876563 | 0.68941 | -0.0736011 | -0.0211699 |
| plaie_tronc | 0.309558 | 0.402056 | -0.382171 | -0.409827 | 0.0126527 | 0.0677316 |
| champignon_houppier | -0.0317644 | -0.329102 | -0.0882225 | 0.511329 | 0.044008 | -0.106249 |
| fissure_houppier | 0.00144877 | 0.282041 | 0.262076 | -0.303087 | -0.243152 | 0.000673708 |
| bois_mort_houppier | 0.78076 | -0.0663505 | -0.449878 | -0.424198 | -0.00514947 | 0.164816 |
| plaie_houppier | 0.509451 | 0.388228 | -0.352651 | -0.455034 | -0.185651 | 0.0956575 |
| esperance_maintien | -1.80137 | -0.419574 | 0.886968 | 1.21003 | 0.810899 | -0.686958 |
| contrainte | -0.0987119 | -0.16551 | -0.0705857 | -0.0418059 | 0.769036 | -0.392422 |
| date_diagnostic | -0.73924 | -0.592679 | 0.805366 | -0.023401 | 0.372522 | 0.177432 |
| prescription_1 | -0.0438974 | 0.347947 | 0.427542 | -0.152804 | -0.836765 | 0.257978 |
| prescription_2 | 0.356812 | 0.198394 | -0.635027 | 0.0896253 | 0.0341129 | -0.0439177 |
| Long | -0.624515 | 0.89516 | -0.374953 | 0.206672 | 0.118174 | -0.220537 |
| Lat | 0.246282 | -0.176627 | 0.681353 | -0.326147 | -0.0619319 | -0.362929 |